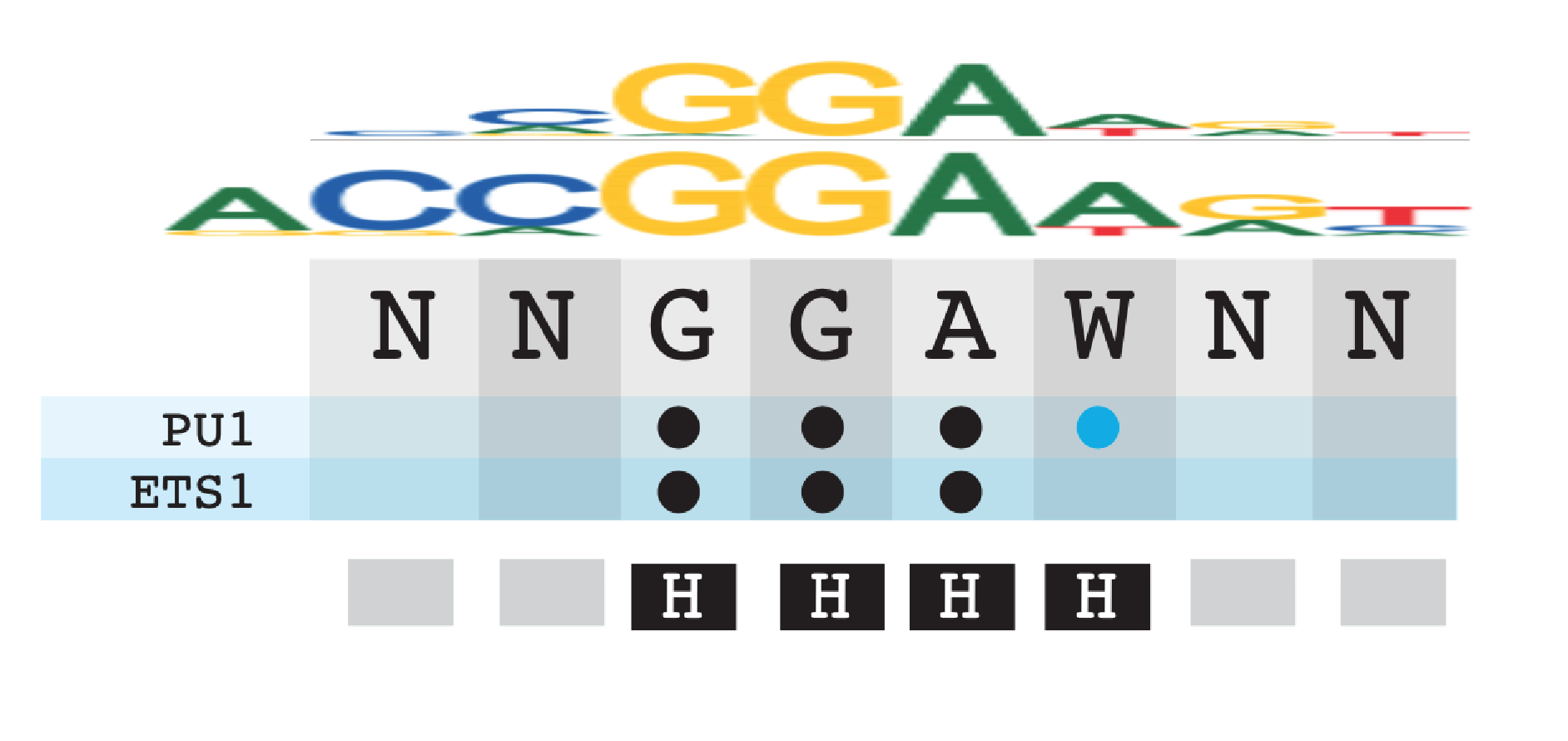
1. Pre-determined TF Core binding sites
2. Summary for attaining core binding site from crystal structure
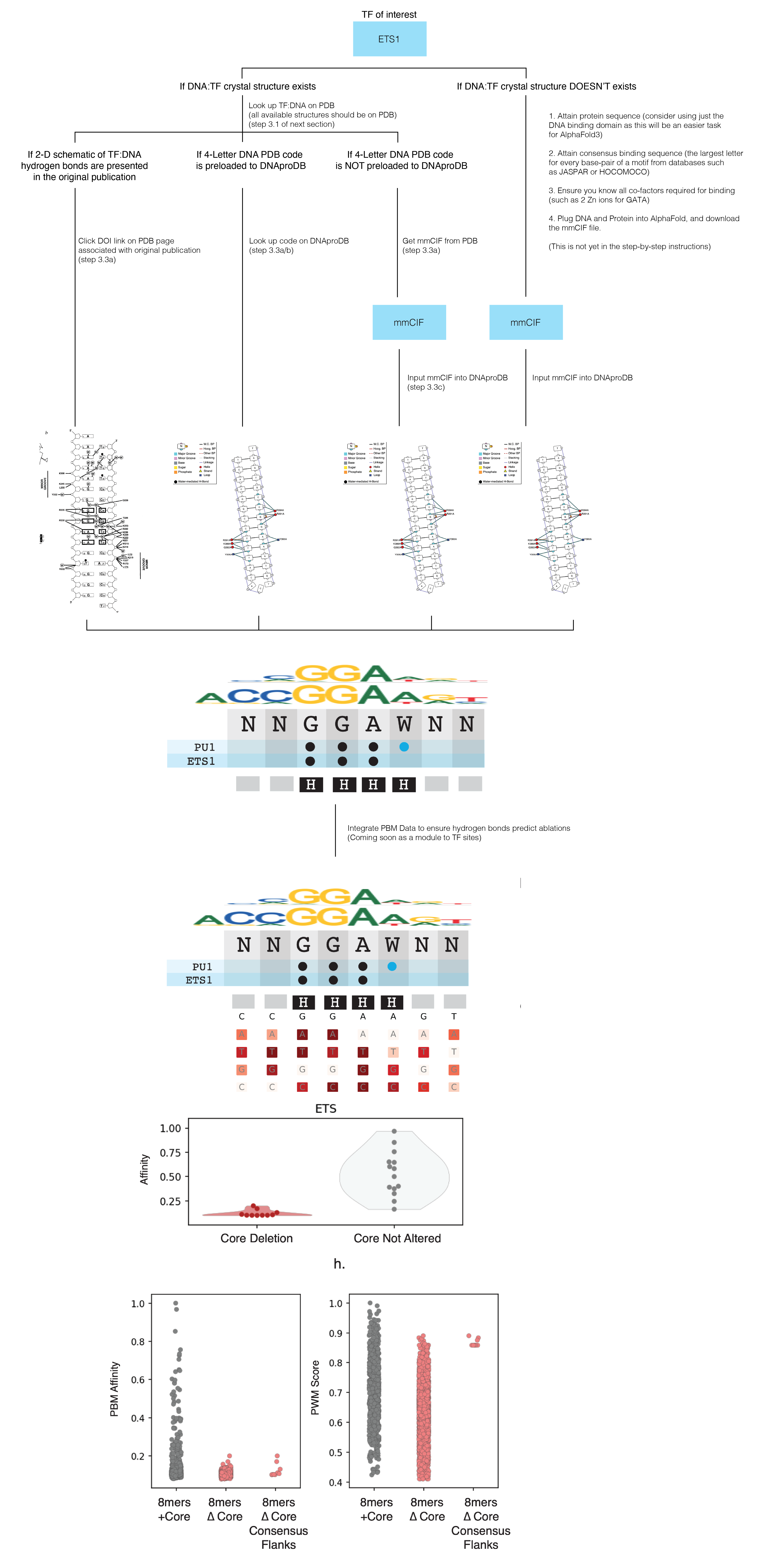
3. Mapping the hydrogen bonds between TF and DNA (step-by-step)
Hydrogen bonds are the strongest kind of interaction between TF and DNA, and typically flag the bases that have the largest contribution to binding affinity. Below describe the steps to determine the bases that make direct H-bonds between the TF and the base of DNA (not the sugar or posphate backbone).
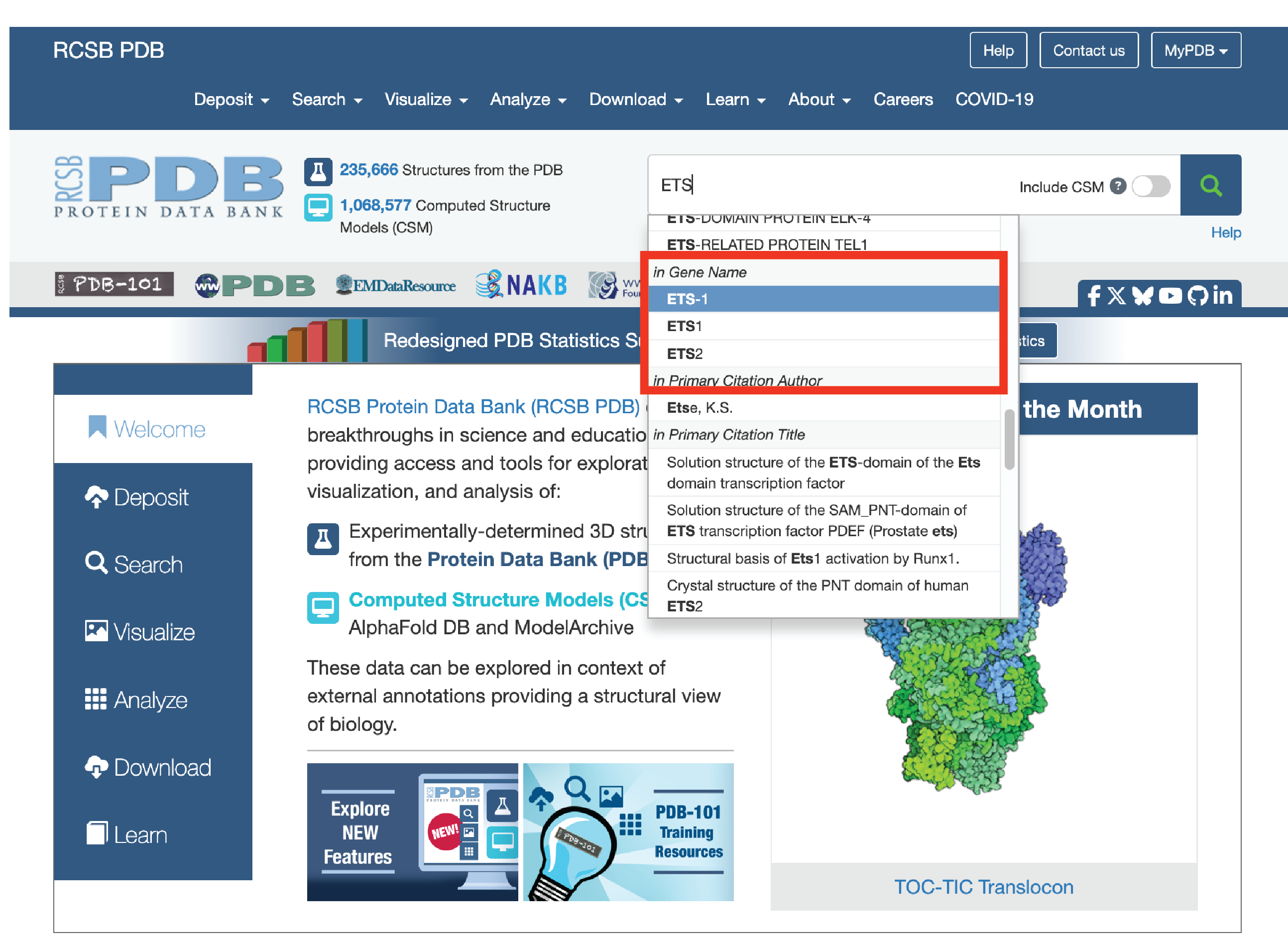
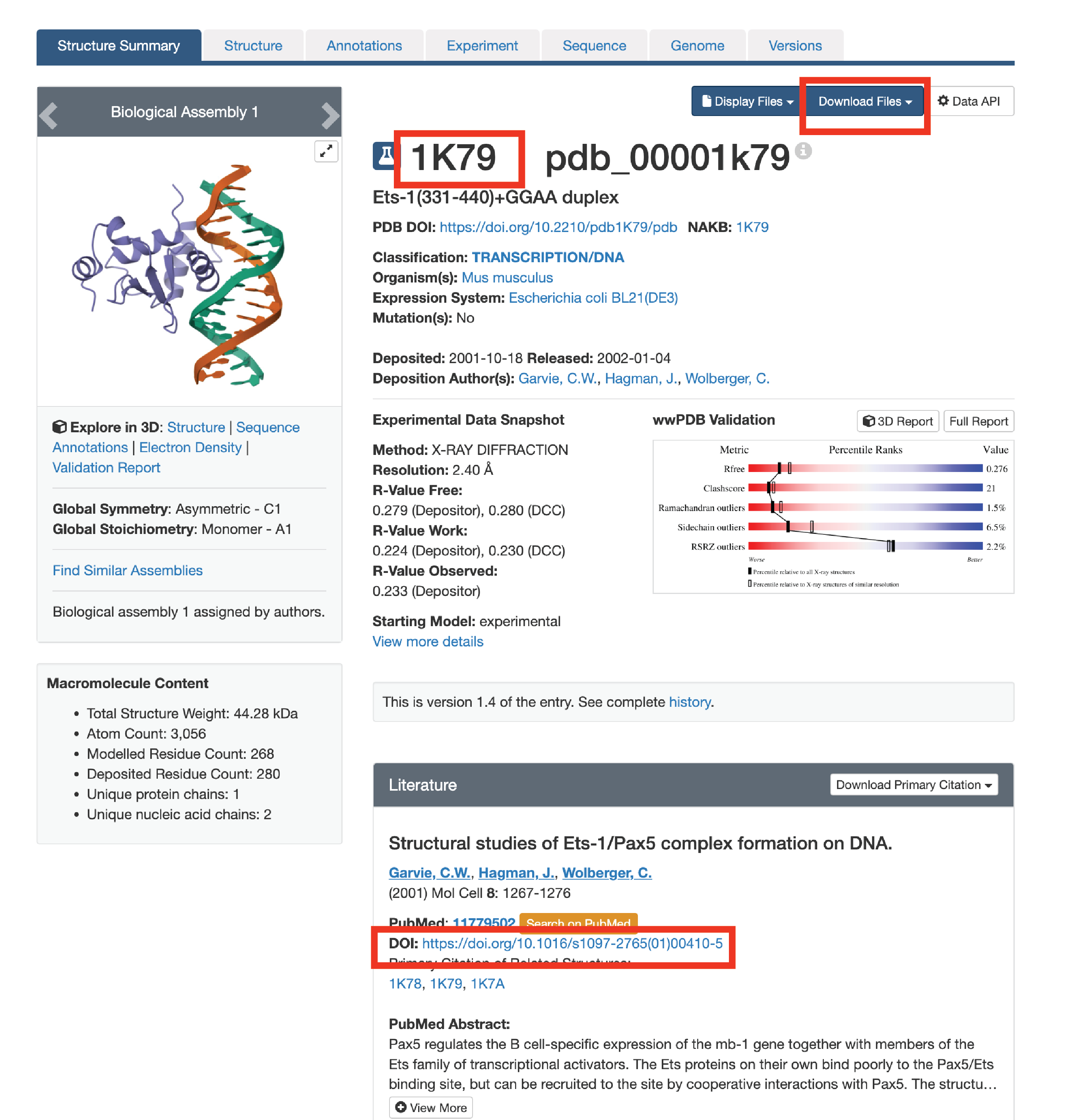
3.1. Attain PDB code or crystal structure (www.rcsb.org). We like to scroll to the “in Gene Name” section of the search function.
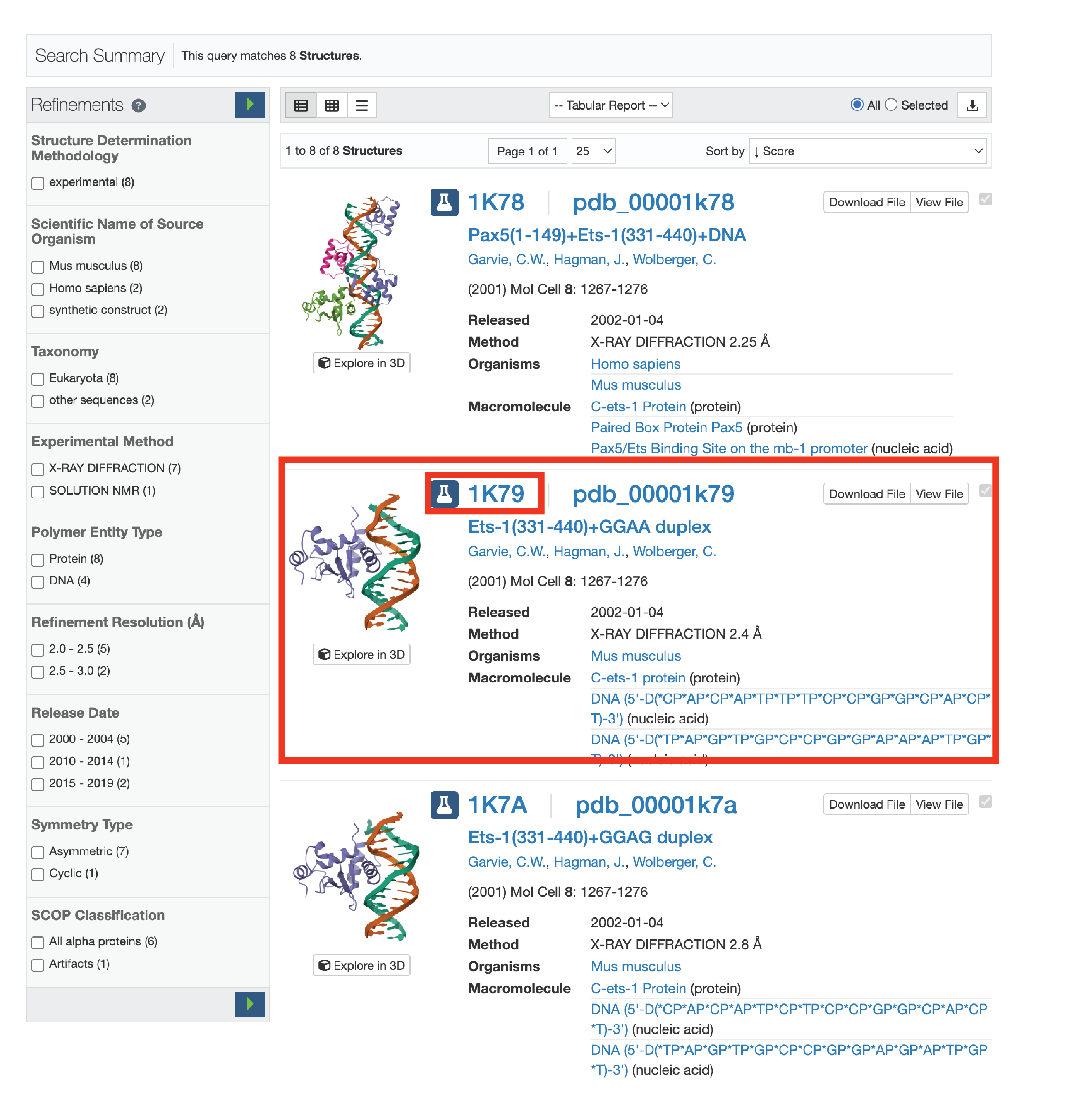
3.2. Locate the structure you want. If there are multiple, it can be helpful to analyze all of them to see what H-bonds are shared. Make sure the crystal structure is the TF-DNA complex. Take note of the 4-letter PDB code.
3.3a. Check if the publication has the 2-D cartoon schematic explaining the H-bonds. If its not available, either note the 4-letter PDB code or download the mmCIF file
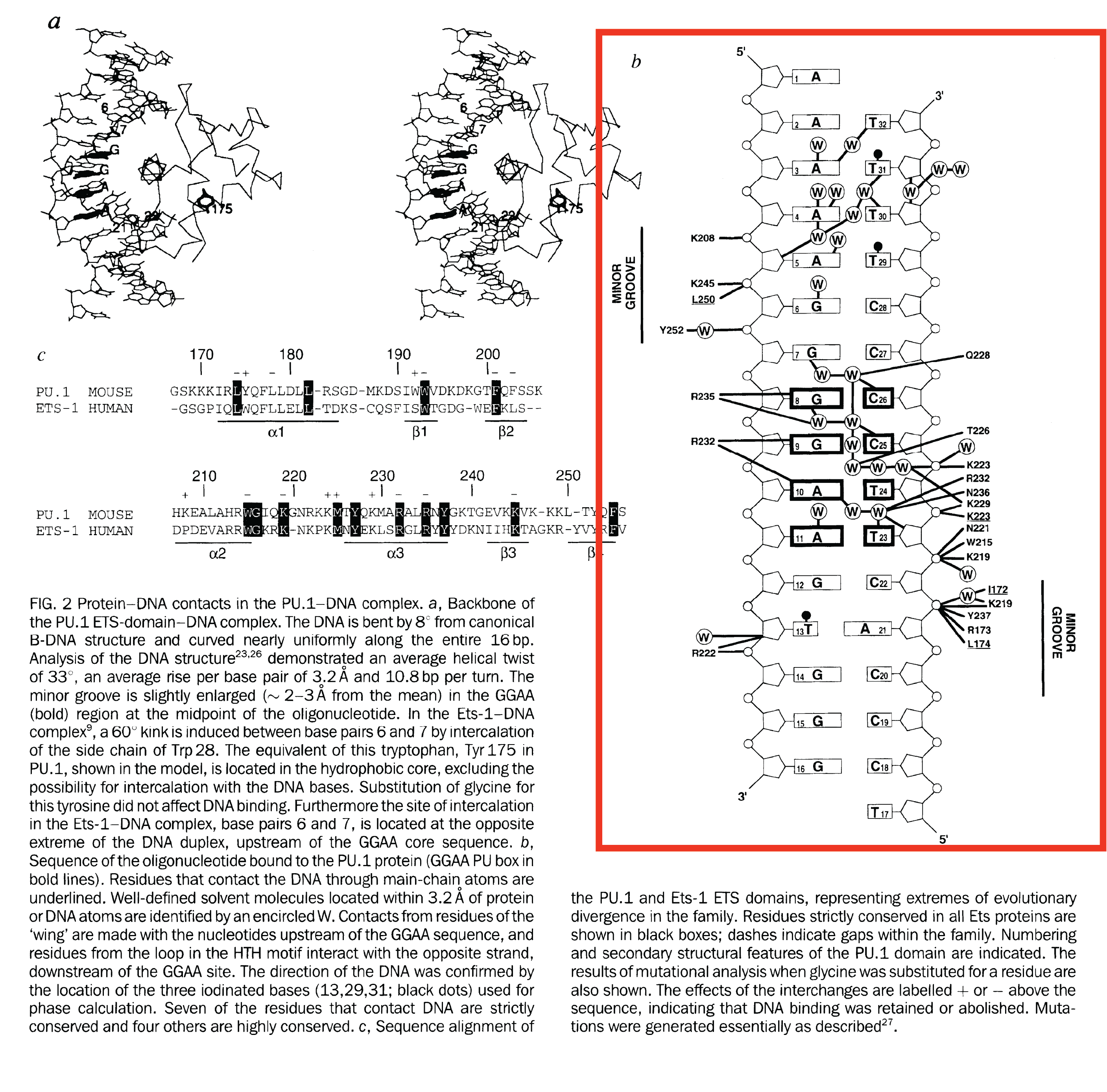
Below is the crystal structure of PU.1 ETS protein (1PUE) to show an example of what the 2-D schematic displaying the H-bonds between TF and DNA.
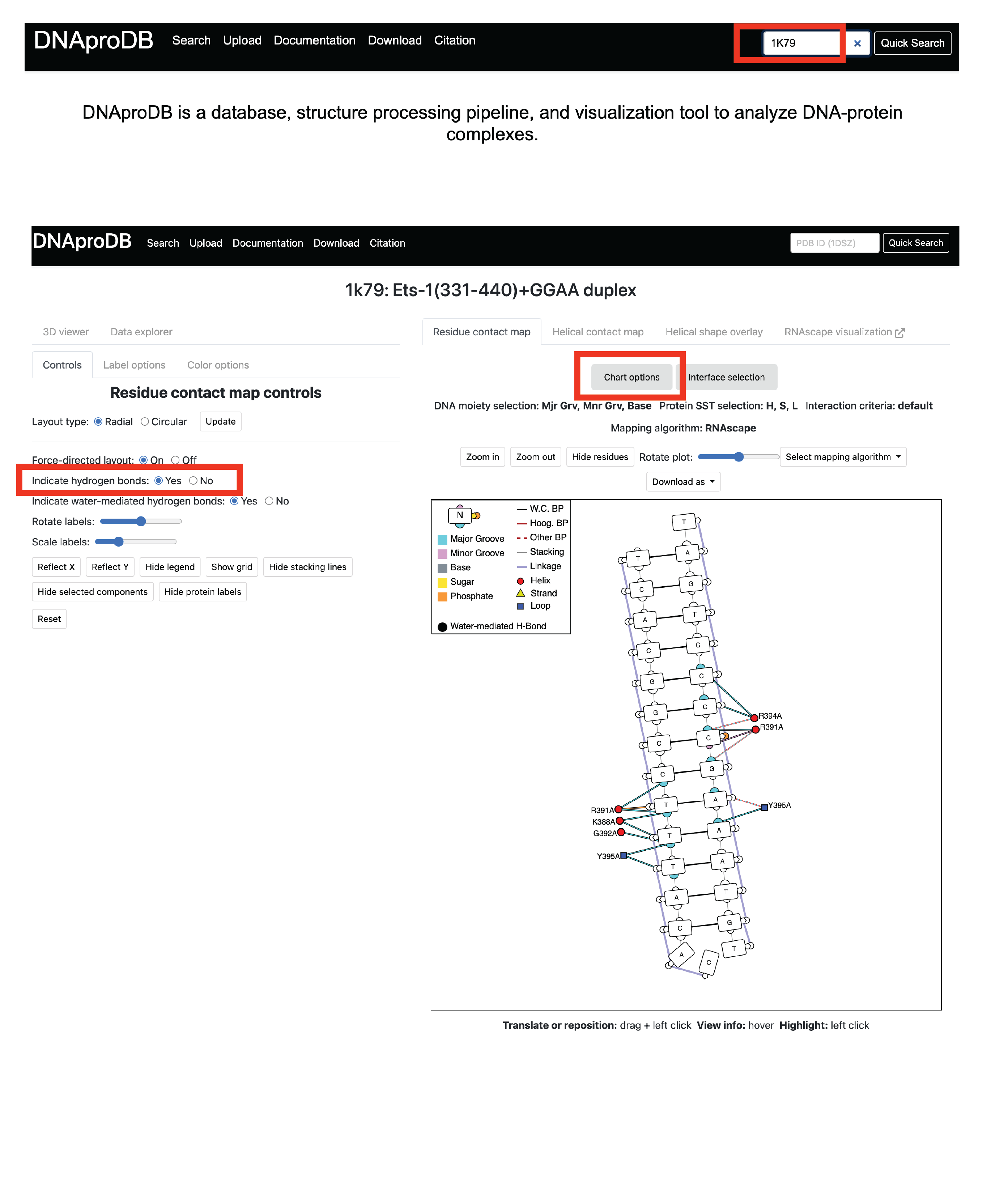
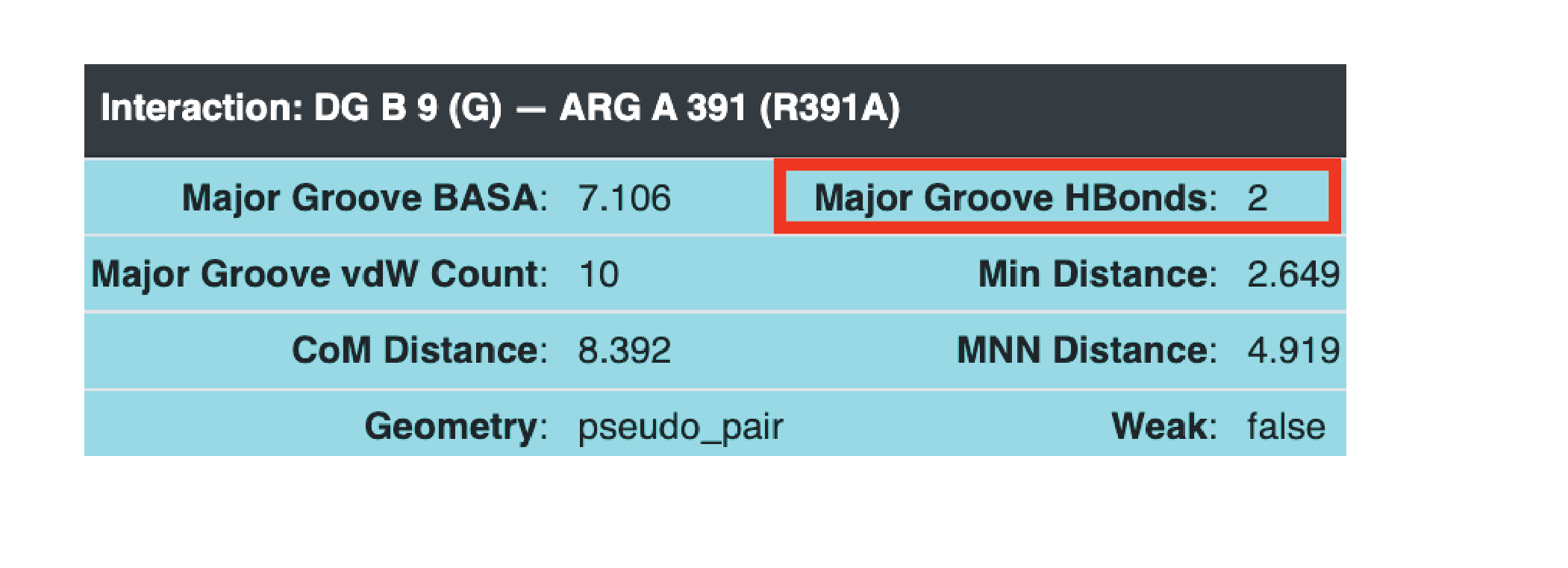
Hover over the edges connecting the amino acid with the nucleotide (top is major groove, bottom is minor groove). Below is an example of a amino acid hydrogen bonding with the major groove base. In this case no amino acids hydrogen bond with the minor groove, but if they were they would show up with lines that contact the other side of the base.
3.3c. Upload mmCIF downloaded from PDB to DNAproDB
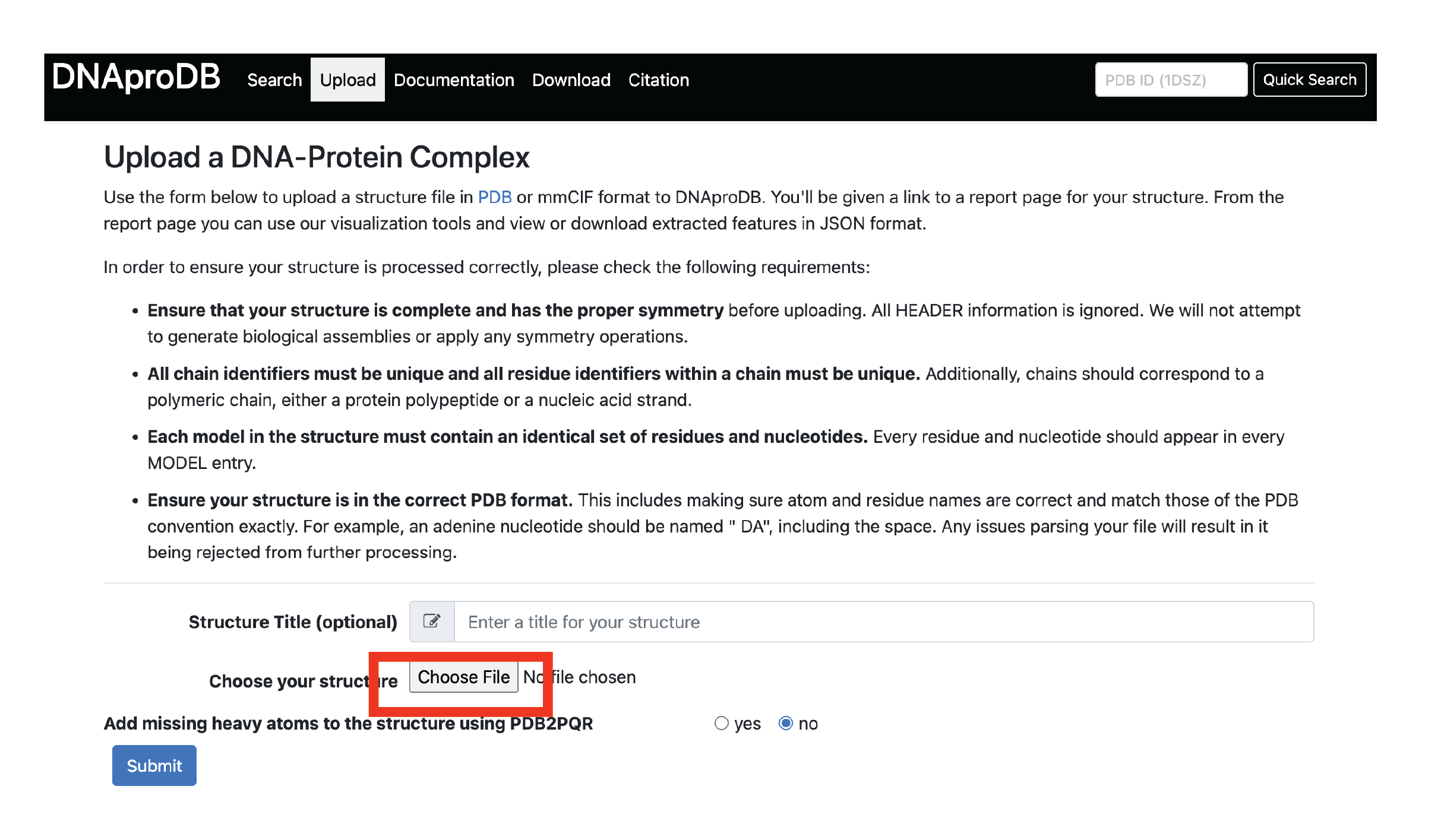
3.4. Curate all information using PWMs and the hydrogen bonds.